https://doi.org/10.1140/epje/s10189-025-00500-8
Regular Article - Living Systems
A python-based novel vertex–edge-weighted modeling framework for enhanced QSPR analysis of cardiovascular and diabetes drug molecules
1
Department of Mathematics, Nevşehir Hacı Bektaş Veli University, 50300, Nevşehir, Turkey
2
Department of Mathematical Sciences, Karakoram International University Gilgit, Gilgit, Pakistan
Received:
30
April
2025
Accepted:
10
June
2025
Published online:
8
July
2025
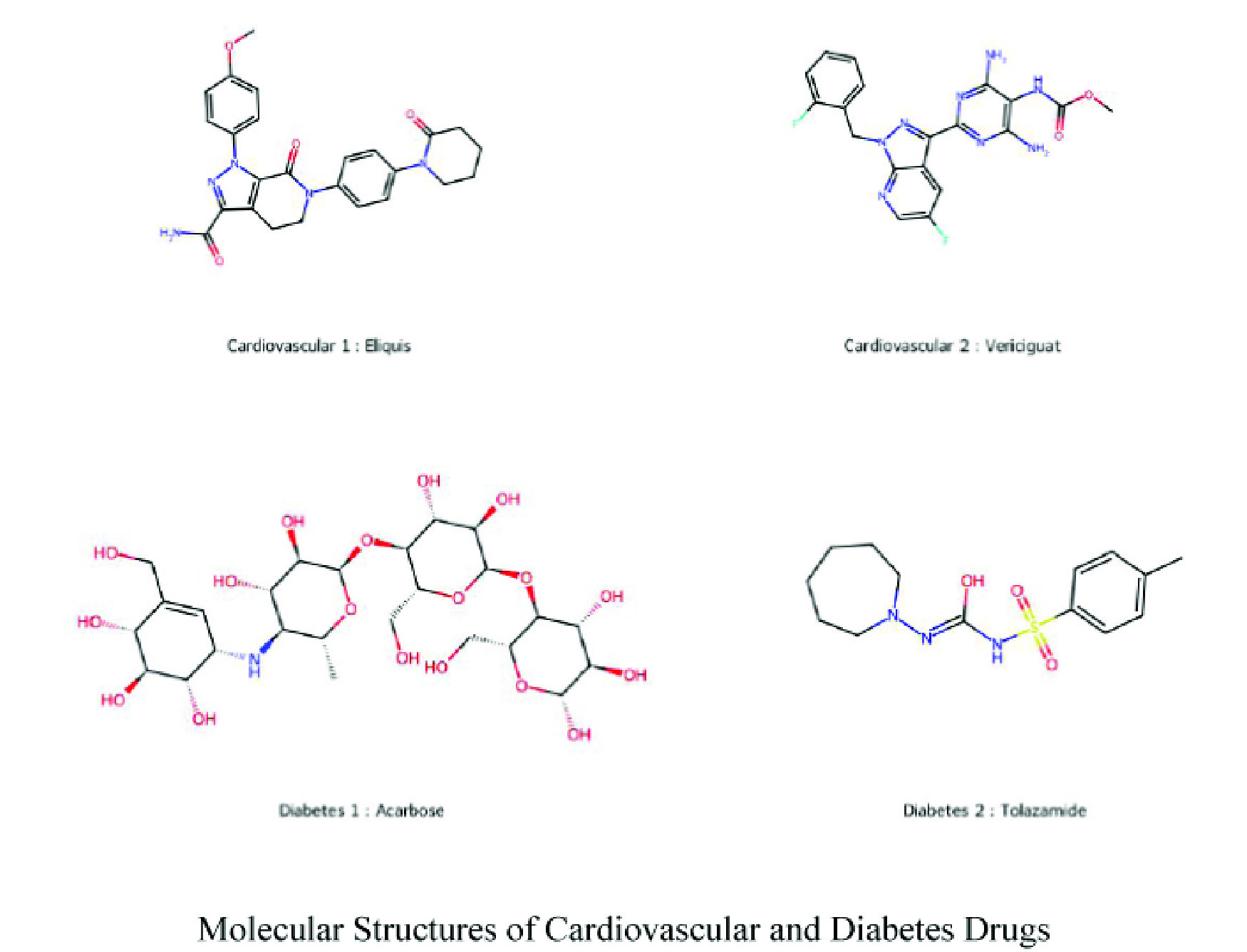
This study advances the quantitative structure–property relationship analysis by leveraging novel vertex–edge-weighted (VEW) molecular graphs to investigate 19 drug molecules commonly used to treat cardiovascular diseases and diabetes. The graphs are constructed by assigning weights to vertices and edges based on atomic properties, enabling a detailed and chemically meaningful representation of molecular structures. Python-based programs were developed to compute degree-based topological indices, which were then analyzed through robust linear regression models to uncover correlations with key physicochemical properties. The results reveal strong and consistent relationships between the computed indices and the physicochemical properties, validating the predictive capability of the proposed approach. Notably, the VEW model demonstrates significant improvements in accuracy and correlation strength over traditional unweighted molecular graph models, underscoring its enhanced ability to capture intricate molecular interactions. This work provides novel insights into the utility of degree-based topological indices in drug design, particularly for cardiovascular and diabetic treatments. By bridging theoretical modeling with practical pharmaceutical applications, it lays a solid foundation for optimizing molecular properties, improving drug efficacy, and accelerating the drug development pipeline. These findings reaffirm the growing significance of computational strategies in advancing precision medicine and pharmaceutical innovation.
Copyright comment Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
© The Author(s), under exclusive licence to EDP Sciences, SIF and Springer-Verlag GmbH Germany, part of Springer Nature 2025
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.