https://doi.org/10.1140/epje/s10189-025-00509-z
Regular Article - Living Systems
Probing protein–protein interactions with drag flow: a case study of F-actin and tropomyosin
1
Université Paris-Cité, CNRS, Institut Jacques Monod, F-75013, Paris, France
2
Université Paris-Saclay, CNRS, CentraleSupélec, Laboratoire EM2C, 91190, Gif-sur-Yvette, France
3
Structural Motility, Institut Curie, Université Paris Sciences et Lettres, Sorbonne Université, CNRS UMR144, 75248, Paris, France
Received:
28
April
2025
Accepted:
9
July
2025
Published online:
13
August
2025
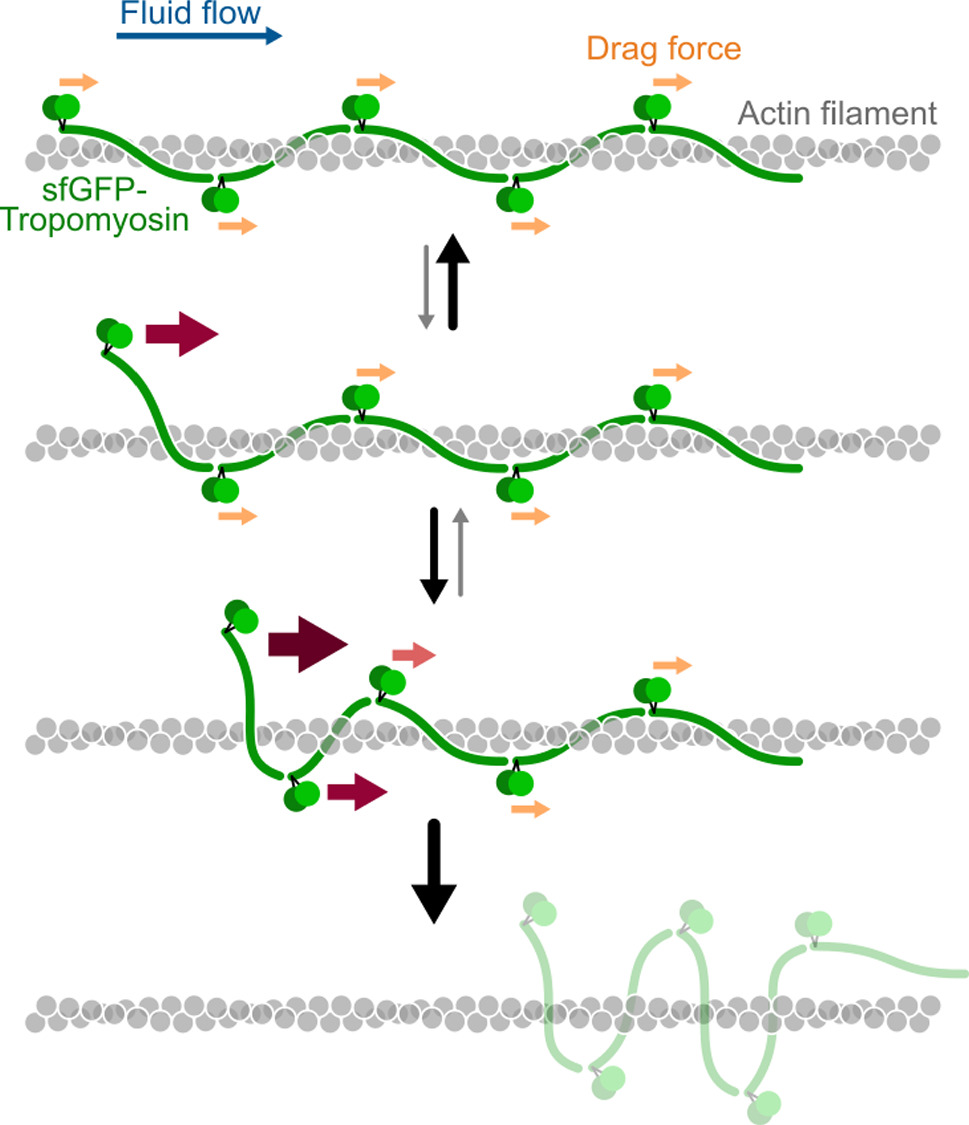
Tropomyosins are central regulators of the actin cytoskeleton, controlling the binding and activity of the other actin binding proteins. The interaction between tropomyosin and actin is quite unique: single tropomyosin dimers bind weakly to actin filaments but get stabilised by end-to-end attachment with neighbouring tropomyosin dimers, forming clusters which wrap around the filament. Force spectroscopy is a powerful approach for studying protein–protein interactions, but classical methods which usually pull with pN forces on a single protein pair, are not well adapted to tropomyosins. Here, we propose a method in which a hydrodynamic drag force is applied directly to the proteins of interest, by imposing a controlled fluid flow inside a microfluidic chamber. The breaking of the protein bonds is directly visualised with fluorescence microscopy. Using this approach, we reveal that very low forces from 0.01 to 0.1 pN per tropomyosin dimer trigger the detachment of entire tropomyosin clusters from actin filaments. We show that the tropomyosin cluster detachment rate depends on the cytoplasmic tropomyosin isoform (Tpm1.6, 1.7, 1.8) and increases exponentially with the applied force. These observations lead us to propose a cluster detachment model which suggests that tropomyosins dynamically explore different positions over the actin filament. Our experimental setup can be used with many other cytoskeletal proteins, and we show, as a proof-of-concept, that the velocity of myosin-X motors is reduced by an opposing fluid flow. Overall, this method expands the range of protein–protein interactions that can be studied by force spectroscopy.
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1140/epje/s10189-025-00509-z.
Copyright comment Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
© The Author(s), under exclusive licence to EDP Sciences, SIF and Springer-Verlag GmbH Germany, part of Springer Nature 2025
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.