https://doi.org/10.1140/epje/s10189-025-00491-6
Regular Article - Living Systems
Improved QSAR methods for predicting drug properties utilizing topological indices and machine learning models
1
College of Mathematical Sciences, Harbin Engineering University, Harbin, People’s Republic of China
2
Department of Computer Science and Information Technology, Women University of Azad Jammu & Kashmir, Bagh, Pakistan
3
School of Mathematical Sciences, Anhui University, 230601, Hefei, People’s Republic of China
4
Department of Mathematics, Women University of Azad Jammu & Kashmir, Bagh, Pakistan
Received:
18
December
2024
Accepted:
7
April
2025
Published online:
9
May
2025
This research investigates the anticipated physicochemical and topological properties of compounds such as drug complexity (C), molecular weight (MW), and topological polar surface area (TPSA) using quantitative structure–activity relationship (QSAR) analysis. Several machine learning models, including Linear Regression, Ridge Regression, Lasso Regression, Random Forest Regression, and Gradient Boosting, were developed to improve prediction accuracy using topological indices. The datasets were combined with appropriate topological indices for individual compounds. Model performance was evaluated using Mean Squared Error (MSE) and 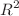 score after hyperparameter tuning via GridSearchCV. Ridge and Lasso Regression models stood out due to their lowest Test MSE averages (3617.74 and 3540.23, respectively) and highest
score after hyperparameter tuning via GridSearchCV. Ridge and Lasso Regression models stood out due to their lowest Test MSE averages (3617.74 and 3540.23, respectively) and highest 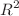 scores (0.9322 and 0.9374, respectively), demonstrating their effectiveness in handling multicollinearity and preventing overfitting. Linear Regression also performed robustly, achieving an MSE of 5249.97 and an
scores (0.9322 and 0.9374, respectively), demonstrating their effectiveness in handling multicollinearity and preventing overfitting. Linear Regression also performed robustly, achieving an MSE of 5249.97 and an 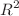 of 0.8563, highlighting the suitability of simpler models for datasets with inherent linear relationships. While Random Forest and Gradient Boosting Regression are capable of capturing nonlinear relationships, their performance varied. Random Forest Regression achieved an MSE of 6485.45 and an
of 0.8563, highlighting the suitability of simpler models for datasets with inherent linear relationships. While Random Forest and Gradient Boosting Regression are capable of capturing nonlinear relationships, their performance varied. Random Forest Regression achieved an MSE of 6485.45 and an 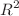 of 0.6643, while Gradient Boosting initially performed poorly with an MSE of 4488.04 and an
of 0.6643, while Gradient Boosting initially performed poorly with an MSE of 4488.04 and an 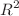 of 0.5659. After fine-tuning Gradient Boosting with an expanded hyperparameter grid, its performance improved significantly, achieving a Test MSE of 1494.74 and an
of 0.5659. After fine-tuning Gradient Boosting with an expanded hyperparameter grid, its performance improved significantly, achieving a Test MSE of 1494.74 and an 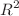 of 0.9171. However, it still ranked fourth, suggesting that simpler models like Linear, Ridge, and Lasso Regression may be better suited for this dataset. This work emphasizes the significance of accurate model selection and optimization in QSAR analysis, demonstrating how these approaches can be used to develop dependable predictive models in computational drug discovery and cheminformatics.
of 0.9171. However, it still ranked fourth, suggesting that simpler models like Linear, Ridge, and Lasso Regression may be better suited for this dataset. This work emphasizes the significance of accurate model selection and optimization in QSAR analysis, demonstrating how these approaches can be used to develop dependable predictive models in computational drug discovery and cheminformatics.
Copyright comment Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
© The Author(s), under exclusive licence to EDP Sciences, SIF and Springer-Verlag GmbH Germany, part of Springer Nature 2025
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.





