https://doi.org/10.1140/epje/i2019-11920-x
Regular Article
A novel combinatorial approach of quantitative microscopy and in silico modeling deciphers Arf1-dependent Golgi size regulation
1
Department of Cell and Tumor Biology, Advanced Centre for Treatment Research & Education in Cancer (ACTREC), Tata Memorial Centre, Plot No. 1 & 2, Sector 22, Kharghar, 410210, Navi Mumbai, Maharashtra, India
2
Homi Bhabha National Institute, Training School Complex, Anushakti Nagar, MH 400085, Mumbai, India
3
Department of Solid State Physics, Indian Association for the Cultivation of Science, 700032, Jadavpur, Kolkata, India
* e-mail: ssprp@iacs.res.in
** e-mail: dbhattacharyya@actrec.gov.in
Received:
23
July
2019
Accepted:
21
November
2019
Published online:
12
December
2019
Regulation of organelle size and shape is a poorly understood but fascinating subject. Several theoretical studies were reported on Golgi size regulation, but a combination of experimental and theoretical approaches is rare. In combination with the quantitative microscopy and a coarse-grained simulation model, we have developed a technique to gain insights into the functions of potential regulators of Golgi size in budding yeast Saccharomyces cerevisiae. To validate our method, we tested wild-type and arf1
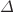 strain harboring early and late Golgi cisternae labeled with green and red fluorescent fusions. Our concentration-dependent maturation model prediction concurs with most of the experimental results for both wild-type and arf1
strain harboring early and late Golgi cisternae labeled with green and red fluorescent fusions. Our concentration-dependent maturation model prediction concurs with most of the experimental results for both wild-type and arf1
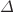 strains. Decisive match of simulation and experimental data provide insight into such specific factor's function in regulating the Golgi size. Details of the complex multifactorial network of Golgi size regulation can be deciphered in the future using a similar combination of quantitative microscopy and in silico model.
strains. Decisive match of simulation and experimental data provide insight into such specific factor's function in regulating the Golgi size. Details of the complex multifactorial network of Golgi size regulation can be deciphered in the future using a similar combination of quantitative microscopy and in silico model.
Key words: Living systems: Cellular Processes
© EDP Sciences / Società Italiana di Fisica / Springer-Verlag GmbH Germany, part of Springer Nature, 2019





